Tutorial 6. Modify trees and states
Summary:
This tutorial will take you through the process of generating random trees, editing trees and states, converting states from continuous to discrete, and combining results.
Information of data source
The consensus tree and 100 trees were downloaded from the 10k Trees Website (https://10ktrees.nunn-lab.org, Arnold et al., 2010). The sociality character is modified from Shultz et al. (2011). Other characters are modified from Kamilar and Cooper (2013).
All files used in this tutorial are stored in examples/Primate/. If you are a beginner of RASP, please start from Tutorial 1.
Loading the data files
Click [File> Close Current Data] or reopen RASP to clear the current trees. We will use a different trees dataset in this tutorial.
Open [File > Load Trees> Load Trees (more format)] and navigate to Trees_States /100Trees.trees.
Open [File> Load Consensus Tree> Load User-specified Tree] and navigate to Trees_States/Primates.tree.
Open [File > Load States (Distributions)], navigate to Trees_States/characters.csv.
Add species to trees dataset
Suppose you want to add a species to the most recent common ancestor of Theropithecus geladag and Papio hamadryas (node 43)
Open [Tools> Add Omitted Groups], you will see a window like this:
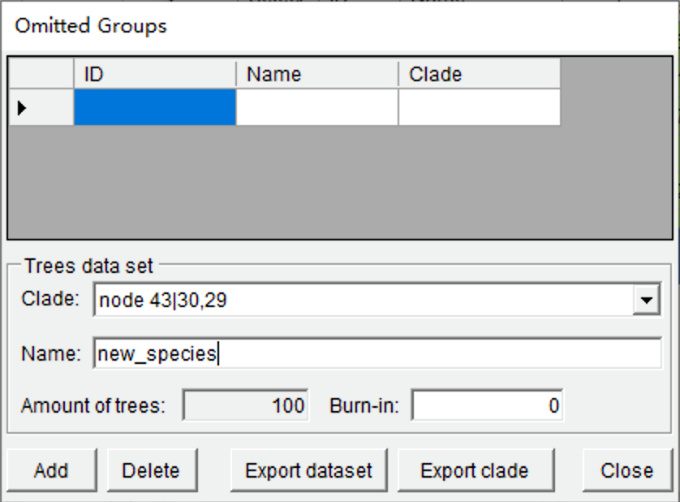
Select node 43 and give the name of the new species (new_species) here. Click the “Add” button and then click the “Export dataset” button to save the modified tree.
NOTE: You will get a tree containing the new species without branch lengths.
Remove species from your data
Suppose you want to remove Eulemur fulvus and Lemur catta from the trees and states. Click the select box in the head of the two species. You will see a window like this:
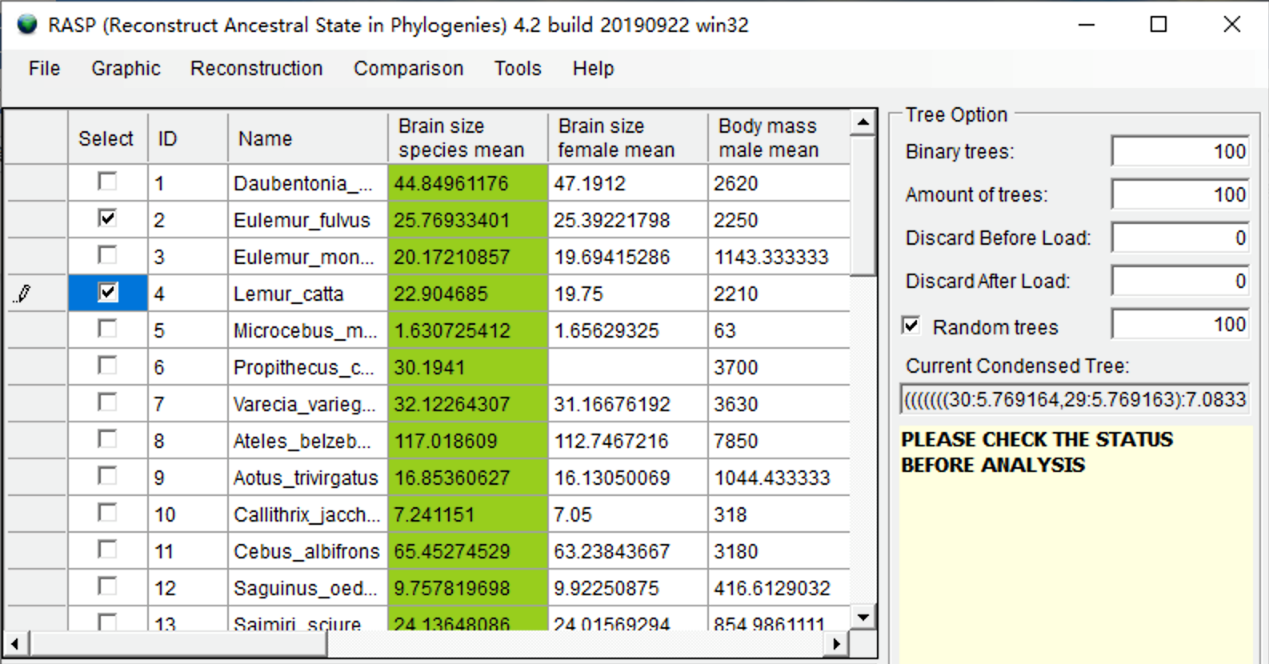
Open [Tools> Remove Selected Groups] and save the new data to a folder. You will get a new consensus tree, trees dataset and states file without your selected species. The branch lengths of the trees will be kept.
Scaling branch lengths
Scaling branch lengths may be useful if you want to change the unit of absolute time of your trees from millions of years to ten thousand years, or if some of the branch lengths are too small to analyze using the BGB method. You can scale branch lengths using RASP by opening [Tools> Scaling Branch Length].
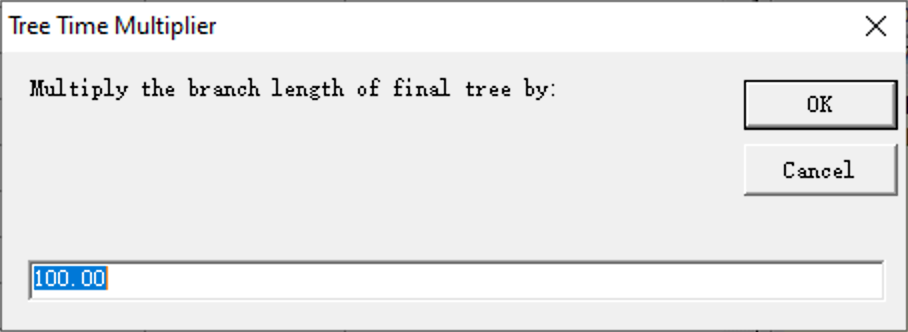
Enter a float number in input box, and the branch lengths of the consensus tree will be multiplied by this value.
Generate and save random trees
In many methods implemented in RASP, we use the “random trees” option for quick analysis. If you want to repeat the analysis exactly, you can set a seed for the random trees to obtain the same “random trees” for different analyses. If you do not set this number, the seed will be generated using the time of your system. Open [Tools> Seed For Random Trees] to set the seed. Open [File> Export Trees> Random Trees] to save the random trees.
Convert states from continuous to discrete
Select the “Brain size species mean” character by clicking the header of the column. Open [Tools> Edit States> Convert States] and you will see a window like this:

Change the number at the bottom to set the number of partitions. Users can also change the range values in the table. Click the Apply button when ready. You will be asked to give a new name to the state. The new state will be added to the end of the state table.
Combine the results
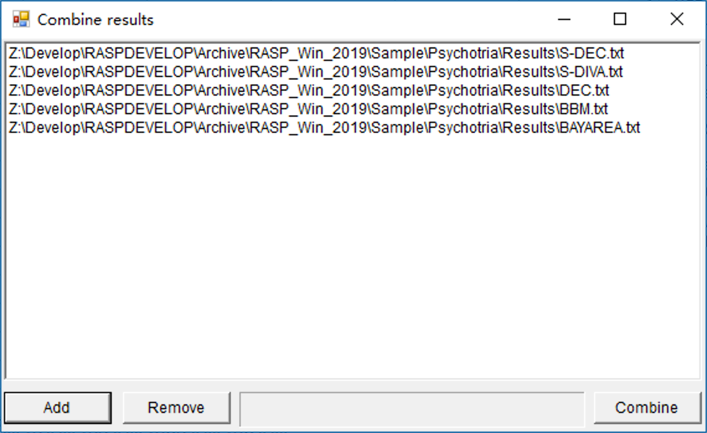
RASP results generated from the same dataset may be combined. If you get multiple results using different methods or run a method (like BayArea) several times to get several results, you can combine them to obtain a single result.
Open [Tools > Combine Results] to combine and save different results.
References
Arnold, C., Matthews, L. J., & Nunn, C. L. (2010). The 10kTrees website: a new online resource for primate phylogeny. Evolutionary Anthropology: Issues, News, and Reviews, 19(3), 114-118.
Kamilar, J. M., & Cooper, N. (2013). Phylogenetic signal in primate behaviour, ecology and life history. Philos Trans R Soc Lond B Biol Sci, 368(1618), 20120341.
Shultz, S., Opie, C., & Atkinson, Q. D. (2011). Stepwise evolution of stable sociality in primates. Nature, 479(7372), 219-222.